class: center, middle, inverse, title-slide # Joins ### Daniel Anderson ### Week 3, Class 2 --- layout: true <script> feather.replace() </script> <div class="slides-footer"> <span> <a class = "footer-icon-link" href = "https://github.com/uo-datasci-specialization/c2-dataviz-2021/raw/main/static/slides/w3p2.pdf"> <i class = "footer-icon" data-feather="download"></i> </a> <a class = "footer-icon-link" href = "https://dataviz-2021.netlify.app/slides/w3p2.html"> <i class = "footer-icon" data-feather="link"></i> </a> <a class = "footer-icon-link" href = "https://github.com/uo-datasci-specialization/c2-dataviz-2021"> <i class = "footer-icon" data-feather="github"></i> </a> </span> </div> --- # Learning Objectives * Understand and be able to identify keys * Understand different types of joins + left, right, inner, full + one-to-one, one-to-many * Understand common ways joins fail * Understand the difference between mutating and filtering joins --- # Before we get started * Today we'll talk about both **mutating** and **filtering** joins * Mutating joins are more common, but filtering joins can be really powerful * Mutating joins add columns to a dataset -- ### What if I want to add rows? * Not technically a join (no key involved, which we'll talk about momentarily) --- # Quick example, binding rows ```r g3 <- tibble(sid = 1:3, grade = rep(3, 3), score = as.integer(rnorm(3, 200,10))) g4 <- tibble(sid = 9:11, grade = rep(4, 3), score = as.integer(rnorm(3, 200,10))) ``` .pull-left[ ```r g3 ``` ``` ## # A tibble: 3 x 3 ## sid grade score ## <int> <dbl> <int> ## 1 1 3 203 ## 2 2 3 180 ## 3 3 3 191 ``` ] .pull-left[ ```r g4 ``` ``` ## # A tibble: 3 x 3 ## sid grade score ## <int> <dbl> <int> ## 1 9 4 213 ## 2 10 4 206 ## 3 11 4 210 ``` ] --- # `bind_rows` * In examples like the previous datasets, we just want to "staple" the rows together. * We can do so with `bind_rows`. ```r bind_rows(g3, g4) ``` ``` ## # A tibble: 6 x 3 ## sid grade score ## <int> <dbl> <int> ## 1 1 3 203 ## 2 2 3 180 ## 3 3 3 191 ## 4 9 4 213 ## 5 10 4 206 ## 6 11 4 210 ``` --- # Optional `.id` argument * What if we knew the grade, but didn't have a variable in each dataset already? * Use `.id` to add an index for each dataset ```r bind_rows(g3[ ,-2], g4[ ,-2], .id = "dataset") ``` ``` ## # A tibble: 6 x 3 ## dataset sid score ## <chr> <int> <int> ## 1 1 1 203 ## 2 1 2 180 ## 3 1 3 191 ## 4 2 9 213 ## 5 2 10 206 ## 6 2 11 210 ``` --- ```r bind_rows(g3[ ,-2], g4[ ,-2], .id = "dataset") %>% mutate(grade = ifelse(dataset == 1, 3, 4)) ``` ``` ## # A tibble: 6 x 4 ## dataset sid score grade ## <chr> <int> <int> <dbl> ## 1 1 1 203 3 ## 2 1 2 180 3 ## 3 1 3 191 3 ## 4 2 9 213 4 ## 5 2 10 206 4 ## 6 2 11 210 4 ``` --- # Even better usage ```r bind_rows(g3 = g3[ ,-2], g4 = g4[ ,-2], .id = "grade") ``` ``` ## # A tibble: 6 x 3 ## grade sid score ## <chr> <int> <int> ## 1 g3 1 203 ## 2 g3 2 180 ## 3 g3 3 191 ## 4 g4 9 213 ## 5 g4 10 206 ## 6 g4 11 210 ``` --- ## What if columns don't match exactly? Pad with `NA` ```r bind_rows(g3, g4[ ,-2], .id = "dataset") ``` ``` ## # A tibble: 6 x 4 ## dataset sid grade score ## <chr> <int> <dbl> <int> ## 1 1 1 3 203 ## 2 1 2 3 180 ## 3 1 3 3 191 ## 4 2 9 NA 213 ## 5 2 10 NA 206 ## 6 2 11 NA 210 ``` --- # Last note - read in a bunch of files * We'll talk about this a lot more in the next course * `purrr::map_df` uses `bind_rows` in the background -- ```r dir.create("tmp") mtcars %>% split(.$cyl) %>% walk2(c("tmp/cyl4.csv", "tmp/cyl6.csv", "tmp/cyl8.csv"), write_csv) list.files("tmp") ``` ``` ## [1] "cyl4.csv" "cyl6.csv" "cyl8.csv" ``` --- # Read in files Use `purrr::map_df` with the file names Note `fs::dir_ls` is equivalent to `list.files`, but plays nicer with `purrr::map_df` ```r new_mtcars <- map_df(fs::dir_ls("tmp"), rio::import, setclass = "tbl_df", .id = "file") new_mtcars %>% select(file, mpg, cyl) %>% slice(1:3) ``` ``` ## # A tibble: 3 x 3 ## file mpg cyl ## <chr> <dbl> <int> ## 1 tmp/cyl4.csv 22.8 4 ## 2 tmp/cyl4.csv 24.4 4 ## 3 tmp/cyl4.csv 22.8 4 ``` ```r unlink("tmp", recursive = TRUE) ``` --- class: inverse-blue center middle # Joins (not to be confused with row binding) --- # Keys * Uniquely identify rows in a dataset -- * Variable(s) in common between two datasets to be joined -- + A key can be more than one variable -- ### Types of keys * Small distinction that you probably won't have to worry about much, but is worth mentioning: + **Primary keys:** Uniquely identify observations in their dataset + **Foreign keys:** Uniquely identify observations in other datasets. --- # What's the primary key here? ```r library(rio) library(here) ecls <- import(here("data", "ecls-k_samp.sav"), setclass = "tbl_df") %>% characterize() ecls ``` ``` ## # A tibble: 984 x 33 ## child_id teacher_id school_id k_type school_type sex ## <chr> <chr> <chr> <chr> <chr> <chr> ## 1 0842021C 0842T02 0842 full-day public male ## 2 0905002C 0905T01 0905 full-day private male ## 3 0150012C 0150T01 0150 full-day private female ## 4 0556009C 0556T01 0556 full-day private female ## 5 0089013C 0089T04 0089 full-day public male ## 6 1217001C 1217T13 1217 half-day public female ## 7 1092008C 1092T01 1092 half-day public female ## 8 0083007C 0083T16 0083 full-day public male ## 9 1091005C 1091T02 1091 half-day private male ## 10 2006006C 2006T01 2006 full-day private male ## # … with 974 more rows, and 27 more variables: ethnic <chr>, famtype <chr>, ## # numsibs <dbl>, SES_cont <dbl>, SES_cat <chr>, age <dbl>, T1RSCALE <dbl>, ## # T1MSCALE <dbl>, T1GSCALE <dbl>, T2RSCALE <dbl>, T2MSCALE <dbl>, ## # T2GSCALE <dbl>, IRTreadgain <dbl>, IRTmathgain <dbl>, IRTgkgain <dbl>, ## # T1ARSLIT <dbl>, T1ARSMAT <dbl>, T1ARSGEN <dbl>, T2ARSLIT <dbl>, ## # T2ARSMAT <dbl>, T2ARSGEN <dbl>, ARSlitgain <dbl>, ARSmathgain <dbl>, ## # ARSgkgain <dbl>, testdate1 <date>, testdate2 <date>, elapse <dbl> ``` --- # Double-checking ```r ecls %>% count(child_id) ``` ``` ## # A tibble: 984 x 2 ## child_id n ## * <chr> <int> ## 1 0001010C 1 ## 2 0002010C 1 ## 3 0009005C 1 ## 4 0009014C 1 ## 5 0009026C 1 ## 6 0013003C 1 ## 7 0016004C 1 ## 8 0016009C 1 ## 9 0022005C 1 ## 10 0022014C 1 ## # … with 974 more rows ``` --- ```r ecls %>% count(child_id) %>% filter(n > 1) ``` ``` ## # A tibble: 0 x 2 ## # … with 2 variables: child_id <chr>, n <int> ``` --- # What about here? ```r income_ineq <- read_csv(here("data", "incomeInequality_tidy.csv")) print(income_ineq, n = 15) ``` ``` ## # A tibble: 726 x 6 ## Year Number.thousands realGDPperCap PopulationK percentile income ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 1947 37237 14117.32 144126 20 14243 ## 2 1947 37237 14117.32 144126 40 22984 ## 3 1947 37237 14117.32 144126 60 31166 ## 4 1947 37237 14117.32 144126 80 44223 ## 5 1947 37237 14117.32 144126 50 26764.14 ## 6 1947 37237 14117.32 144126 90 41477 ## 7 1947 37237 14117.32 144126 95 54172 ## 8 1947 37237 14117.32 144126 99 134415 ## 9 1947 37237 14117.32 144126 99.5 203001 ## 10 1947 37237 14117.32 144126 99.9 479022 ## 11 1947 37237 14117.32 144126 99.99 1584506 ## 12 1948 38624 14451.94 146631 20 13779 ## 13 1948 38624 14451.94 146631 40 22655 ## 14 1948 38624 14451.94 146631 60 30248 ## 15 1948 38624 14451.94 146631 80 42196 ## # … with 711 more rows ``` --- ```r income_ineq %>% count(Year, percentile) %>% filter(n > 1) ``` ``` ## # A tibble: 0 x 3 ## # … with 3 variables: Year <dbl>, percentile <dbl>, n <int> ``` --- # Sometimes there is no key These tables have an *implicit* id - the row numbers. For example: ```r install.packages("nycflights13") library(nycflights13) ``` ```r head(flights) ``` ``` ## # A tibble: 6 x 19 ## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time ## <int> <int> <int> <int> <int> <dbl> <int> <int> ## 1 2013 1 1 517 515 2 830 819 ## 2 2013 1 1 533 529 4 850 830 ## 3 2013 1 1 542 540 2 923 850 ## 4 2013 1 1 544 545 -1 1004 1022 ## 5 2013 1 1 554 600 -6 812 837 ## 6 2013 1 1 554 558 -4 740 728 ## # … with 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>, ## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>, ## # hour <dbl>, minute <dbl>, time_hour <dttm> ``` --- ```r flights %>% count(year, month, day, flight, tailnum) %>% filter(n > 1) ``` ``` ## # A tibble: 11 x 6 ## year month day flight tailnum n ## <int> <int> <int> <int> <chr> <int> ## 1 2013 2 9 303 <NA> 2 ## 2 2013 2 9 655 <NA> 2 ## 3 2013 2 9 1623 <NA> 2 ## 4 2013 6 8 2269 N487WN 2 ## 5 2013 6 15 2269 N230WN 2 ## 6 2013 6 22 2269 N440LV 2 ## 7 2013 6 29 2269 N707SA 2 ## 8 2013 7 6 2269 N259WN 2 ## 9 2013 8 3 2269 N446WN 2 ## 10 2013 8 10 2269 N478WN 2 ## 11 2013 12 15 398 <NA> 2 ``` --- # Create a key * If there is no key, it's often helpful to add one. These are called *surrogate* keys. ```r flights <- flights %>% rowid_to_column() flights %>% select(1:3, ncol(flights)) ``` ``` ## # A tibble: 336,776 x 4 ## rowid year month time_hour ## <int> <int> <int> <dttm> ## 1 1 2013 1 2013-01-01 05:00:00 ## 2 2 2013 1 2013-01-01 05:00:00 ## 3 3 2013 1 2013-01-01 05:00:00 ## 4 4 2013 1 2013-01-01 05:00:00 ## 5 5 2013 1 2013-01-01 06:00:00 ## 6 6 2013 1 2013-01-01 05:00:00 ## 7 7 2013 1 2013-01-01 06:00:00 ## 8 8 2013 1 2013-01-01 06:00:00 ## 9 9 2013 1 2013-01-01 06:00:00 ## 10 10 2013 1 2013-01-01 06:00:00 ## # … with 336,766 more rows ``` --- class: inverse-red middle # Mutating joins --- # Mutating joins * In *tidyverse*, we use `mutate()` to create new variables within a dataset. -- * A mutating join works similarly, in that we're adding new variables to the existing dataset through a join. -- * Two tables of data joined by a common key --- # Four types of joins * `left_join`: Keep all the data in the left dataset, drop any non-matching cases from the right dataset. * `right_join`: Keep all the data in the right dataset, drop any non-matching cases from the left dataset. * `inner_join`: Keep only data that matches in both datasets * `full_join`: Keep all the data in both datasets. This is also sometimes referred to as an *outer* join. -- If the keys match exactly in the two tables (datasets), all of these will result in the .bolder[exact] same result. --- # Using joins to recode Say you have a dataset like this ```r set.seed(1) disab_codes <- c("00", "10", "20", "40", "43", "50", "60", "70", "74", "80", "82", "90", "96", "98") dis_tbl <- tibble( sid = 1:200, dis_code = sample(disab_codes, 200, replace = TRUE), score = as.integer(rnorm(200, 200, 10)) ) head(dis_tbl) ``` ``` ## # A tibble: 6 x 3 ## sid dis_code score ## <int> <chr> <int> ## 1 1 74 190 ## 2 2 40 200 ## 3 3 60 200 ## 4 4 00 183 ## 5 5 10 210 ## 6 6 96 188 ``` --- # Codes <br/> .pull-left[ | Code | Disability| |:----:|:----------| | 00 |'Not Applicable' | | 10 |'Intellectual Disability' | | 20 |'Hearing Impairment'| | 40 |'Visual Impairment'| | 43 |'Deaf-Blindness'| | 50 |'Communication Disorder'| | 60 |'Emotional Disturbance'| | 70 |'Orthopedic Impairment'| | 74 |'Traumatic Brain Injury'| ] .pull-right[ | Code | Disability| |:----:|:----------| | 80 |'Other Health Impairments'| | 82 |'Autism Spectrum Disorder'| | 90 |'Specific Learning Disability'| | 96 |'Developmental Delay 0-2yr'| | 98 |'Developmental Delay 3-4yr'| ] --- # One method ```r dis_tbl %>% mutate(disability = case_when( dis_code == "10" ~ "Intellectual Disability", dis_code == "20" ~ 'Hearing Impairment', ..., TRUE ~ "Not Applicable" ) ) ``` --- # Joining method ```r dis_code_tbl <- tibble( dis_code = c( "00", "10", "20", "40", "43", "50", "60", "70", "74", "80", "82", "90", "96", "98" ), disability = c( 'Not Applicable', 'Intellectual Disability', 'Hearing Impairment', 'Visual Impairment', 'Deaf-Blindness', 'Communication Disorder', 'Emotional Disturbance', 'Orthopedic Impairment', 'Traumatic Brain Injury', 'Other Health Impairments', 'Autism Spectrum Disorder', 'Specific Learning Disability', 'Developmental Delay 0-2yr', 'Developmental Delay 3-4yr' ) ) ``` --- ```r dis_code_tbl ``` ``` ## # A tibble: 14 x 2 ## dis_code disability ## <chr> <chr> ## 1 00 Not Applicable ## 2 10 Intellectual Disability ## 3 20 Hearing Impairment ## 4 40 Visual Impairment ## 5 43 Deaf-Blindness ## 6 50 Communication Disorder ## 7 60 Emotional Disturbance ## 8 70 Orthopedic Impairment ## 9 74 Traumatic Brain Injury ## 10 80 Other Health Impairments ## 11 82 Autism Spectrum Disorder ## 12 90 Specific Learning Disability ## 13 96 Developmental Delay 0-2yr ## 14 98 Developmental Delay 3-4yr ``` --- # Join the tables ```r left_join(dis_tbl, dis_code_tbl) ``` ``` ## Joining, by = "dis_code" ``` ``` ## # A tibble: 200 x 4 ## sid dis_code score disability ## <int> <chr> <int> <chr> ## 1 1 74 190 Traumatic Brain Injury ## 2 2 40 200 Visual Impairment ## 3 3 60 200 Emotional Disturbance ## 4 4 00 183 Not Applicable ## 5 5 10 210 Intellectual Disability ## 6 6 96 188 Developmental Delay 0-2yr ## 7 7 60 203 Emotional Disturbance ## 8 8 82 204 Autism Spectrum Disorder ## 9 9 98 201 Developmental Delay 3-4yr ## 10 10 10 198 Intellectual Disability ## # … with 190 more rows ``` --- class: inverse-orange middle # Imperfect key match? --- # Consider the following ```r gender <- tibble(key = 1:3, male = rbinom(3, 1, .5)) sped <- tibble(key = c(1, 2, 4), sped = rbinom(3, 1, .5)) ``` .pull-left[ ```r gender ``` ``` ## # A tibble: 3 x 2 ## key male ## <int> <int> ## 1 1 0 ## 2 2 1 ## 3 3 0 ``` ] .pull-right[ ```r sped ``` ``` ## # A tibble: 3 x 2 ## key sped ## <dbl> <int> ## 1 1 0 ## 2 2 1 ## 3 4 0 ``` ] --- # `left_join()`? -- ```r left_join(gender, sped) ``` ``` ## # A tibble: 3 x 3 ## key male sped ## <dbl> <int> <int> ## 1 1 0 0 ## 2 2 1 1 ## 3 3 0 NA ``` -- # `right_join()`? -- ```r right_join(gender, sped) ``` ``` ## # A tibble: 3 x 3 ## key male sped ## <dbl> <int> <int> ## 1 1 0 0 ## 2 2 1 1 ## 3 4 NA 0 ``` --- # `inner_join()`? -- ```r inner_join(gender, sped) ``` ``` ## # A tibble: 2 x 3 ## key male sped ## <dbl> <int> <int> ## 1 1 0 0 ## 2 2 1 1 ``` -- # `full_join()`? -- ```r full_join(gender, sped) ``` ``` ## # A tibble: 4 x 3 ## key male sped ## <dbl> <int> <int> ## 1 1 0 0 ## 2 2 1 1 ## 3 3 0 NA ## 4 4 NA 0 ``` --- background-image:url(img/joins.png) background-size:contain .footnote[From [r4ds](https://r4ds.had.co.nz)] --- class: inverse-red middle # Animations All of the following animations were created by Garrick Aden-Buie and can be found [here](https://github.com/gadenbuie/tidyexplain) --- # Animated `left_join()`  --- # Animated `right_join`  --- # Animated `inner_join`  --- # Animated `full_join`  --- # What if the key is not unique? * Not a problem, as long as they are unique in .bolder[one] of the tables. + In this case, it's called a one-to-many join <br/> <div align = "center"> <img src = img/one_to_many.png width = 1000> </div> --- # Animated one-to-many join  --- # Example A dataset with school IDs ```r stu <- tibble( sid = 1:9, scid = c(1, 1, 1, 1, 2, 2, 3, 3, 3), score = c(10, 12, 15, 8, 9, 11, 12, 15, 17) ) stu ``` ``` ## # A tibble: 9 x 3 ## sid scid score ## <int> <dbl> <dbl> ## 1 1 1 10 ## 2 2 1 12 ## 3 3 1 15 ## 4 4 1 8 ## 5 5 2 9 ## 6 6 2 11 ## 7 7 3 12 ## 8 8 3 15 ## 9 9 3 17 ``` --- A school-level dataset ```r schl <- tibble( scid = 1:3, stu_tch_ratio = c(22.05, 31.14, 24.87), per_pupil_spending = c(15741.08, 11732.24, 13027.88) ) schl ``` ``` ## # A tibble: 3 x 3 ## scid stu_tch_ratio per_pupil_spending ## <int> <dbl> <dbl> ## 1 1 22.05 15741.08 ## 2 2 31.14 11732.24 ## 3 3 24.87 13027.88 ``` --- # One to many ```r left_join(stu, schl) ``` ``` ## # A tibble: 9 x 5 ## sid scid score stu_tch_ratio per_pupil_spending ## <int> <dbl> <dbl> <dbl> <dbl> ## 1 1 1 10 22.05 15741.08 ## 2 2 1 12 22.05 15741.08 ## 3 3 1 15 22.05 15741.08 ## 4 4 1 8 22.05 15741.08 ## 5 5 2 9 31.14 11732.24 ## 6 6 2 11 31.14 11732.24 ## 7 7 3 12 24.87 13027.88 ## 8 8 3 15 24.87 13027.88 ## 9 9 3 17 24.87 13027.88 ``` --- ## What if key is not unique to either table? Generally this is an error Result is probably not going to be what you want (cartesian product). <div align = "center"> <img src = img/cartesian_product.png width = 1000> </div> --- # Example ```r seasonal_means <- tibble( scid = rep(1:3, each = 3), season = rep(c("fall", "winter", "spring"), 3), mean = rnorm(3*3) ) seasonal_means ``` ``` ## # A tibble: 9 x 3 ## scid season mean ## <int> <chr> <dbl> ## 1 1 fall 0.3447951 ## 2 1 winter 1.539648 ## 3 1 spring -0.3295142 ## 4 2 fall 0.9483894 ## 5 2 winter -0.4792556 ## 6 2 spring -1.514887 ## 7 3 fall 0.4345367 ## 8 3 winter -0.5195367 ## 9 3 spring -0.8345590 ``` --- ```r left_join(stu, seasonal_means) ``` ``` ## # A tibble: 27 x 5 ## sid scid score season mean ## <int> <dbl> <dbl> <chr> <dbl> ## 1 1 1 10 fall 0.3447951 ## 2 1 1 10 winter 1.539648 ## 3 1 1 10 spring -0.3295142 ## 4 2 1 12 fall 0.3447951 ## 5 2 1 12 winter 1.539648 ## 6 2 1 12 spring -0.3295142 ## 7 3 1 15 fall 0.3447951 ## 8 3 1 15 winter 1.539648 ## 9 3 1 15 spring -0.3295142 ## 10 4 1 8 fall 0.3447951 ## # … with 17 more rows ``` --- # How do we fix this? --  In some cases, the solution is obvious. In others, it's not. But **you must have at least one unique key** to join the datasets. --- # In this case Move the dataset to wide before joining -- ### Move to wide ```r seasonal_means_wide <- seasonal_means %>% pivot_wider(names_from = "season", values_from = "mean") seasonal_means_wide ``` ``` ## # A tibble: 3 x 4 ## scid fall winter spring ## <int> <dbl> <dbl> <dbl> ## 1 1 0.3447951 1.539648 -0.3295142 ## 2 2 0.9483894 -0.4792556 -1.514887 ## 3 3 0.4345367 -0.5195367 -0.8345590 ``` --- # Join One to many join ```r left_join(stu, seasonal_means_wide) ``` ``` ## # A tibble: 9 x 6 ## sid scid score fall winter spring ## <int> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 1 1 10 0.3447951 1.539648 -0.3295142 ## 2 2 1 12 0.3447951 1.539648 -0.3295142 ## 3 3 1 15 0.3447951 1.539648 -0.3295142 ## 4 4 1 8 0.3447951 1.539648 -0.3295142 ## 5 5 2 9 0.9483894 -0.4792556 -1.514887 ## 6 6 2 11 0.9483894 -0.4792556 -1.514887 ## 7 7 3 12 0.4345367 -0.5195367 -0.8345590 ## 8 8 3 15 0.4345367 -0.5195367 -0.8345590 ## 9 9 3 17 0.4345367 -0.5195367 -0.8345590 ``` --- # Move longer again? If we did, we'd be exactly where we were with the first join. You could make the argument it *might* make sense here I'd still argue for *this* approach, not the cartesian product approach More systematic, more predictable, and ultimately less error prone --- # Another example * Often you want to add summary info to your dataset. * You can do this easily with by piping arguments -- ### ECLS-K reminder ```r ecls ``` ``` ## # A tibble: 984 x 33 ## child_id teacher_id school_id k_type school_type sex ## <chr> <chr> <chr> <chr> <chr> <chr> ## 1 0842021C 0842T02 0842 full-day public male ## 2 0905002C 0905T01 0905 full-day private male ## 3 0150012C 0150T01 0150 full-day private female ## 4 0556009C 0556T01 0556 full-day private female ## 5 0089013C 0089T04 0089 full-day public male ## 6 1217001C 1217T13 1217 half-day public female ## 7 1092008C 1092T01 1092 half-day public female ## 8 0083007C 0083T16 0083 full-day public male ## 9 1091005C 1091T02 1091 half-day private male ## 10 2006006C 2006T01 2006 full-day private male ## # … with 974 more rows, and 27 more variables: ethnic <chr>, famtype <chr>, ## # numsibs <dbl>, SES_cont <dbl>, SES_cat <chr>, age <dbl>, T1RSCALE <dbl>, ## # T1MSCALE <dbl>, T1GSCALE <dbl>, T2RSCALE <dbl>, T2MSCALE <dbl>, ## # T2GSCALE <dbl>, IRTreadgain <dbl>, IRTmathgain <dbl>, IRTgkgain <dbl>, ## # T1ARSLIT <dbl>, T1ARSMAT <dbl>, T1ARSGEN <dbl>, T2ARSLIT <dbl>, ## # T2ARSMAT <dbl>, T2ARSGEN <dbl>, ARSlitgain <dbl>, ARSmathgain <dbl>, ## # ARSgkgain <dbl>, testdate1 <date>, testdate2 <date>, elapse <dbl> ``` --- # Compute group means ```r ecls %>% group_by(school_id) %>% summarize(sch_pre_math = mean(T1MSCALE)) ``` ``` ## # A tibble: 515 x 2 ## school_id sch_pre_math ## * <chr> <dbl> ## 1 0001 20.45800 ## 2 0002 14.977 ## 3 0009 18.82 ## 4 0013 42.321 ## 5 0016 17.55100 ## 6 0022 17.8465 ## 7 0023 15.5050 ## 8 0025 19.446 ## 9 0026 16.866 ## 10 0028 14.354 ## # … with 505 more rows ``` --- # Join right within pipeline ```r ecls %>% group_by(school_id) %>% summarize(sch_pre_math = mean(T1MSCALE)) %>% left_join(ecls) %>% select(school_id:k_type) # Just for space ``` ``` ## # A tibble: 984 x 5 ## school_id sch_pre_math child_id teacher_id k_type ## <chr> <dbl> <chr> <chr> <chr> ## 1 0001 20.45800 0001010C 0001T01 full-day ## 2 0002 14.977 0002010C 0002T01 half-day ## 3 0009 18.82 0009026C 0009T01 half-day ## 4 0009 18.82 0009014C 0009T02 half-day ## 5 0009 18.82 0009005C 0009T01 half-day ## 6 0013 42.321 0013003C 0013T01 full-day ## 7 0016 17.55100 0016004C 0016T01 half-day ## 8 0016 17.55100 0016009C 0016T01 half-day ## 9 0022 17.8465 0022005C 0022T01 half-day ## 10 0022 17.8465 0022014C 0022T03 half-day ## # … with 974 more rows ``` --- # Default join behavior By default, the `*_join` functions will use all columns with common names as keys. ```r flights2 <- flights %>% select(year:day, hour, origin, dest, tailnum, carrier) flights2[1:2, ] ``` ``` ## # A tibble: 2 x 8 ## year month day hour origin dest tailnum carrier ## <int> <int> <int> <dbl> <chr> <chr> <chr> <chr> ## 1 2013 1 1 5 EWR IAH N14228 UA ## 2 2013 1 1 5 LGA IAH N24211 UA ``` ```r weather[1:2, ] ``` ``` ## # A tibble: 2 x 15 ## origin year month day hour temp dewp humid wind_dir wind_speed ## <chr> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 EWR 2013 1 1 1 39.02 26.06 59.37 270 10.35702 ## 2 EWR 2013 1 1 2 39.02 26.96 61.63 250 8.05546 ## # … with 5 more variables: wind_gust <dbl>, precip <dbl>, pressure <dbl>, ## # visib <dbl>, time_hour <dttm> ``` --- ```r left_join(flights2, weather) ``` ``` ## Joining, by = c("year", "month", "day", "hour", "origin") ``` ``` ## # A tibble: 336,776 x 18 ## year month day hour origin dest tailnum carrier temp dewp humid ## <int> <int> <int> <dbl> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> ## 1 2013 1 1 5 EWR IAH N14228 UA 39.02 28.04 64.43 ## 2 2013 1 1 5 LGA IAH N24211 UA 39.92 24.98 54.81 ## 3 2013 1 1 5 JFK MIA N619AA AA 39.02 26.96 61.63 ## 4 2013 1 1 5 JFK BQN N804JB B6 39.02 26.96 61.63 ## 5 2013 1 1 6 LGA ATL N668DN DL 39.92 24.98 54.81 ## 6 2013 1 1 5 EWR ORD N39463 UA 39.02 28.04 64.43 ## 7 2013 1 1 6 EWR FLL N516JB B6 37.94 28.04 67.21000 ## 8 2013 1 1 6 LGA IAD N829AS EV 39.92 24.98 54.81 ## 9 2013 1 1 6 JFK MCO N593JB B6 37.94 26.96 64.29 ## 10 2013 1 1 6 LGA ORD N3ALAA AA 39.92 24.98 54.81 ## # … with 336,766 more rows, and 7 more variables: wind_dir <dbl>, ## # wind_speed <dbl>, wind_gust <dbl>, precip <dbl>, pressure <dbl>, ## # visib <dbl>, time_hour <dttm> ``` --- # Use only some vars? If we were joining *flights2* and *planes*, we would not want to use the `year` variable in the join, because **it means different things in each dataset**. ```r head(planes) ``` ``` ## # A tibble: 6 x 9 ## tailnum year type manufacturer model engines ## <chr> <int> <chr> <chr> <chr> <int> ## 1 N10156 2004 Fixed wing multi engine EMBRAER EMB-145XR 2 ## 2 N102UW 1998 Fixed wing multi engine AIRBUS INDUSTRIE A320-214 2 ## 3 N103US 1999 Fixed wing multi engine AIRBUS INDUSTRIE A320-214 2 ## 4 N104UW 1999 Fixed wing multi engine AIRBUS INDUSTRIE A320-214 2 ## 5 N10575 2002 Fixed wing multi engine EMBRAER EMB-145LR 2 ## 6 N105UW 1999 Fixed wing multi engine AIRBUS INDUSTRIE A320-214 2 ## # … with 3 more variables: seats <int>, speed <int>, engine <chr> ``` --- # How? Specify the variables with `by` ```r left_join(flights2, planes, by = "tailnum") ``` ``` ## # A tibble: 336,776 x 16 ## year.x month day hour origin dest tailnum carrier year.y ## <int> <int> <int> <dbl> <chr> <chr> <chr> <chr> <int> ## 1 2013 1 1 5 EWR IAH N14228 UA 1999 ## 2 2013 1 1 5 LGA IAH N24211 UA 1998 ## 3 2013 1 1 5 JFK MIA N619AA AA 1990 ## 4 2013 1 1 5 JFK BQN N804JB B6 2012 ## 5 2013 1 1 6 LGA ATL N668DN DL 1991 ## 6 2013 1 1 5 EWR ORD N39463 UA 2012 ## 7 2013 1 1 6 EWR FLL N516JB B6 2000 ## 8 2013 1 1 6 LGA IAD N829AS EV 1998 ## 9 2013 1 1 6 JFK MCO N593JB B6 2004 ## 10 2013 1 1 6 LGA ORD N3ALAA AA NA ## # … with 336,766 more rows, and 7 more variables: type <chr>, ## # manufacturer <chr>, model <chr>, engines <int>, seats <int>, ## # speed <int>, engine <chr> ``` --- # Mismatched names? * What if you had data to merge like this? ```r names(schl)[1] <- "school_id" schl ``` ``` ## # A tibble: 3 x 3 ## school_id stu_tch_ratio per_pupil_spending ## <int> <dbl> <dbl> ## 1 1 22.05 15741.08 ## 2 2 31.14 11732.24 ## 3 3 24.87 13027.88 ``` ```r stu ``` ``` ## # A tibble: 9 x 3 ## sid scid score ## <int> <dbl> <dbl> ## 1 1 1 10 ## 2 2 1 12 ## 3 3 1 15 ## 4 4 1 8 ## 5 5 2 9 ## 6 6 2 11 ## 7 7 3 12 ## 8 8 3 15 ## 9 9 3 17 ``` --- # Join w/mismatched names ```r left_join(stu, schl, by = c("scid" = "school_id")) ``` ``` ## # A tibble: 9 x 5 ## sid scid score stu_tch_ratio per_pupil_spending ## <int> <dbl> <dbl> <dbl> <dbl> ## 1 1 1 10 22.05 15741.08 ## 2 2 1 12 22.05 15741.08 ## 3 3 1 15 22.05 15741.08 ## 4 4 1 8 22.05 15741.08 ## 5 5 2 9 31.14 11732.24 ## 6 6 2 11 31.14 11732.24 ## 7 7 3 12 24.87 13027.88 ## 8 8 3 15 24.87 13027.88 ## 9 9 3 17 24.87 13027.88 ``` --- class: inverse-blue center middle # filtering joins --- # Filtering joins * `semi_join()` works just like `left_join` or `inner_join` but you don't actually add the variables. * Let's filter classrooms with extremely high math pretest average scores. --- # First, calculate averages ```r av_pre_mth <- ecls %>% mutate(cut_high = mean(T1MSCALE) + 3*sd(T1MSCALE)) %>% group_by(teacher_id, k_type) %>% summarize(av_pre_mth = mean(T1MSCALE), cut_high = unique(cut_high)) av_pre_mth ``` ``` ## # A tibble: 707 x 4 ## # Groups: teacher_id [707] ## teacher_id k_type av_pre_mth cut_high ## <chr> <chr> <dbl> <dbl> ## 1 0001T01 full-day 20.45800 42.62333 ## 2 0002T01 half-day 14.977 42.62333 ## 3 0009T01 half-day 17.6475 42.62333 ## 4 0009T02 half-day 21.165 42.62333 ## 5 0013T01 full-day 42.321 42.62333 ## 6 0016T01 half-day 17.55100 42.62333 ## 7 0022T01 half-day 20.368 42.62333 ## 8 0022T03 half-day 15.325 42.62333 ## 9 0023T01 half-day 10.988 42.62333 ## 10 0023T04 half-day 20.02200 42.62333 ## # … with 697 more rows ``` --- Next, filter for means 3 standard deviations above the mean. ```r extr_high <- av_pre_mth %>% ungroup() %>% filter(av_pre_mth > cut_high) extr_high ``` ``` ## # A tibble: 3 x 4 ## teacher_id k_type av_pre_mth cut_high ## <chr> <chr> <dbl> <dbl> ## 1 0078T04 half-day 45.75 42.62333 ## 2 0663T01 full-day 42.8455 42.62333 ## 3 0944T03 half-day 45.371 42.62333 ``` --- Finally, use `semi_join` to show the full data for these cases ```r semi_join(ecls, extr_high) ``` ``` ## # A tibble: 4 x 33 ## child_id teacher_id school_id k_type school_type sex ## <chr> <chr> <chr> <chr> <chr> <chr> ## 1 0944017C 0944T03 0944 half-day private female ## 2 0663006C 0663T01 0663 full-day private male ## 3 0663012C 0663T01 0663 full-day private female ## 4 0078020C 0078T04 0078 half-day public female ## # … with 27 more variables: ethnic <chr>, famtype <chr>, numsibs <dbl>, ## # SES_cont <dbl>, SES_cat <chr>, age <dbl>, T1RSCALE <dbl>, ## # T1MSCALE <dbl>, T1GSCALE <dbl>, T2RSCALE <dbl>, T2MSCALE <dbl>, ## # T2GSCALE <dbl>, IRTreadgain <dbl>, IRTmathgain <dbl>, IRTgkgain <dbl>, ## # T1ARSLIT <dbl>, T1ARSMAT <dbl>, T1ARSGEN <dbl>, T2ARSLIT <dbl>, ## # T2ARSMAT <dbl>, T2ARSGEN <dbl>, ARSlitgain <dbl>, ARSmathgain <dbl>, ## # ARSgkgain <dbl>, testdate1 <date>, testdate2 <date>, elapse <dbl> ``` --- # Filtering joins `anti_join()` does the opposite of `semi_join`, keeping any rows that do **not** match. ```r nrow(ecls) ``` ``` ## [1] 984 ``` ```r extr_low_ecls <- anti_join(ecls, extr_high) nrow(extr_low_ecls) ``` ``` ## [1] 980 ``` --- # Why is this so beneficial? * Sometimes the boolean logic for `filter` can be overly complicated. -- * Instead, create a data frame that has only the groups you want, and `semi_join` it with your original data -- * Alternatively, create a data frame that has all .bolder[but] the values you want. --- class: inverse-red middle # Stop words ### One more quick example This one is probs more realistic --- # Jane Austen Books ```r # install.packages(c("tidytext", "janeaustenr")) library(tidytext) library(janeaustenr) austen_books() ``` ``` ## # A tibble: 73,422 x 2 ## text book ## * <chr> <fct> ## 1 "SENSE AND SENSIBILITY" Sense & Sensibility ## 2 "" Sense & Sensibility ## 3 "by Jane Austen" Sense & Sensibility ## 4 "" Sense & Sensibility ## 5 "(1811)" Sense & Sensibility ## 6 "" Sense & Sensibility ## 7 "" Sense & Sensibility ## 8 "" Sense & Sensibility ## 9 "" Sense & Sensibility ## 10 "CHAPTER 1" Sense & Sensibility ## # … with 73,412 more rows ``` --- # Get words ```r austen_books() %>% unnest_tokens(word, text) ``` ``` ## # A tibble: 725,055 x 2 ## book word ## <fct> <chr> ## 1 Sense & Sensibility sense ## 2 Sense & Sensibility and ## 3 Sense & Sensibility sensibility ## 4 Sense & Sensibility by ## 5 Sense & Sensibility jane ## 6 Sense & Sensibility austen ## 7 Sense & Sensibility 1811 ## 8 Sense & Sensibility chapter ## 9 Sense & Sensibility 1 ## 10 Sense & Sensibility the ## # … with 725,045 more rows ``` --- # Count words ```r austen_books() %>% unnest_tokens(word, text) %>% count(word, sort = TRUE) ``` ``` ## # A tibble: 14,520 x 2 ## word n ## <chr> <int> ## 1 the 26351 ## 2 to 24044 ## 3 and 22515 ## 4 of 21178 ## 5 a 13408 ## 6 her 13055 ## 7 i 12006 ## 8 in 11217 ## 9 was 11204 ## 10 it 10234 ## # … with 14,510 more rows ``` --- # Plot top 15 words ```r austen_books() %>% unnest_tokens(word, text) %>% count(word, sort = TRUE) %>% mutate(word = fct_reorder(word, n)) %>% slice(1:15) %>% ggplot(aes(word, n)) + geom_col() + coord_flip() ``` --- <!-- --> --- # Stop words ```r stop_words ``` ``` ## # A tibble: 1,149 x 2 ## word lexicon ## <chr> <chr> ## 1 a SMART ## 2 a's SMART ## 3 able SMART ## 4 about SMART ## 5 above SMART ## 6 according SMART ## 7 accordingly SMART ## 8 across SMART ## 9 actually SMART ## 10 after SMART ## # … with 1,139 more rows ``` --- # Remove stop words ```r austen_books() %>% unnest_tokens(word, text) %>% * anti_join(stop_words) %>% count(word, sort = TRUE) ``` ``` ## # A tibble: 13,914 x 2 ## word n ## <chr> <int> ## 1 miss 1855 ## 2 time 1337 ## 3 fanny 862 ## 4 dear 822 ## 5 lady 817 ## 6 sir 806 ## 7 day 797 ## 8 emma 787 ## 9 sister 727 ## 10 house 699 ## # … with 13,904 more rows ``` --- 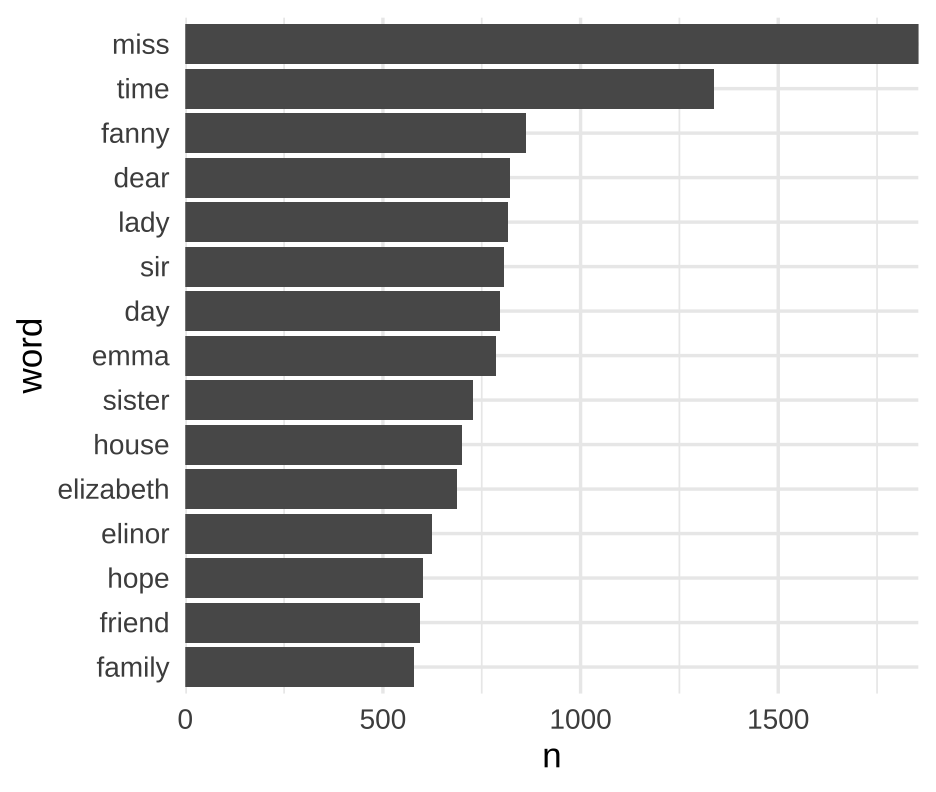<!-- --> --- # By book 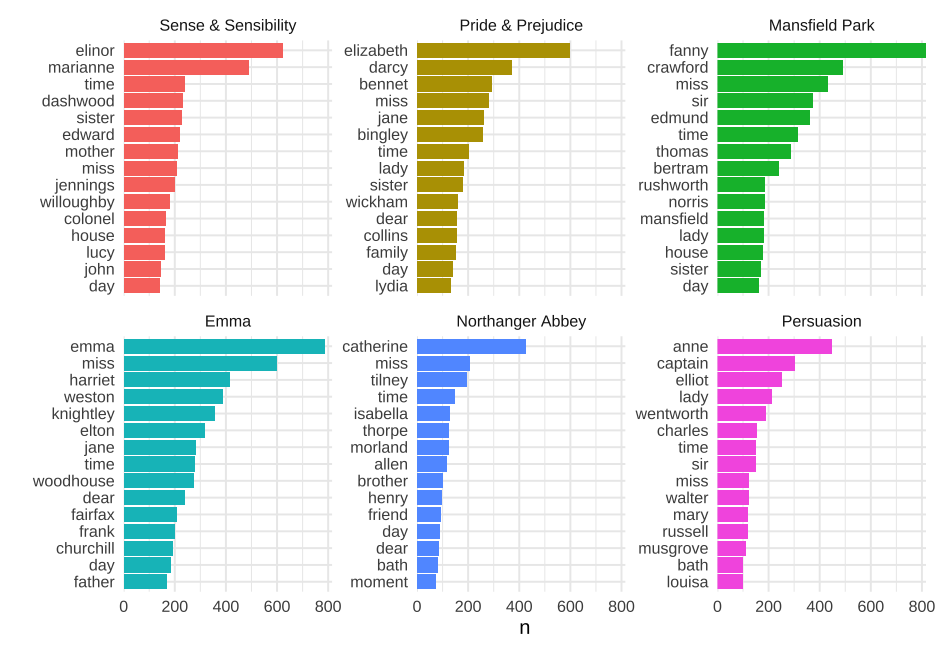<!-- --> --- class: inverse-green middle # Wrapping up * Homework 1 assigned today + Be careful about keys. Likely to be rather tricky. * Next time: Visual perception